Note
Click here to download the full example code or to run this example in your browser via Binder
Unimodal distribution¶
The following example demonstrates the statistical learning based determination of the nuclear shielding tensor parameters from a one-dimensional cross-section of a magic-angle flipping (MAF) spectrum. In this example, we use a synthetic MAF lineshape from a unimodal tensor distribution.
Before getting started¶
Import all relevant packages.
import csdmpy as cp
import matplotlib.pyplot as plt
import numpy as np
from mrinversion.kernel.nmr import ShieldingPALineshape
from mrinversion.linear_model import SmoothLasso, SmoothLassoCV, TSVDCompression
from mrinversion.utils import get_polar_grids
# Setup for the matplotlib figures
# function for 2D x-y plot.
def plot2D(ax, csdm_object, title=""):
# convert the dimension coordinates of the csdm_object from Hz to pmm.
_ = [item.to("ppm", "nmr_frequency_ratio") for item in csdm_object.dimensions]
levels = (np.arange(9) + 1) / 10
plt.figure(figsize=(4.5, 3.5))
ax.contourf(csdm_object, cmap="gist_ncar", levels=levels)
ax.grid(None)
ax.set_title(title)
get_polar_grids(ax)
ax.set_aspect("equal")
Dataset setup¶
Import the dataset¶
Load the dataset. Here, we import the dataset as a CSDM data-object.
# the 1D MAF cross-section data in csdm format
filename = "https://osu.box.com/shared/static/puxfgdh25rru1q3li124anylkgup8rdp.csdf"
data_object = cp.load(filename)
# convert the data dimension from `Hz` to `ppm`.
data_object.dimensions[0].to("ppm", "nmr_frequency_ratio")
The variable data_object holds the 1D MAF cross-section. For comparison, let’s
also import the true tensor parameter distribution from which the synthetic 1D pure
anisotropic MAF cross-section line-shape is simulated.
datafile = "https://osu.box.com/shared/static/s5wpm26w4cv3w64qjhouqu458ch4z0nd.csdf"
true_data_object = cp.load(datafile)
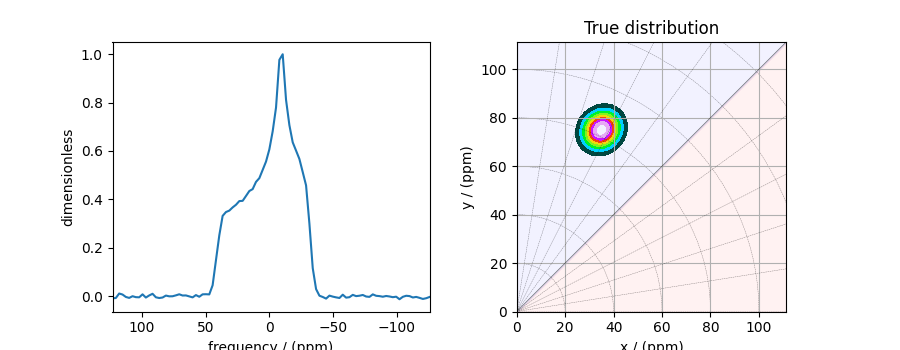
The plot of the 1D MAF cross-section along with the 2D true tensor parameter distribution of the synthetic dataset is shown below.
# the plot of the 1D MAF cross-section dataset.
_, ax = plt.subplots(1, 2, figsize=(9, 3.5), subplot_kw={"projection": "csdm"})
ax[0].plot(data_object)
ax[0].invert_xaxis()
# the plot of the true tensor distribution.
plot2D(ax[1], true_data_object, title="True distribution")
plt.tight_layout()
plt.show()
Linear Inversion setup¶
Dimension setup¶
Anisotropic-dimension: The dimension of the dataset that holds the pure anisotropic frequency contributions, which in this case, is the only dimension.
x-y dimensions: The two inverse dimensions corresponding to the x and y-axis of the x-y grid.
inverse_dimension = [
cp.LinearDimension(count=25, increment="370 Hz", label="x"), # the `x`-dimension.
cp.LinearDimension(count=25, increment="370 Hz", label="y"), # the `y`-dimension.
]
Generating the kernel¶
For MAF datasets, the line-shape kernel corresponds to the pure nuclear shielding
anisotropy line-shapes. Use the
ShieldingPALineshape class to generate a
shielding line-shape kernel.
lineshape = ShieldingPALineshape(
anisotropic_dimension=anisotropic_dimension,
inverse_dimension=inverse_dimension,
channel="29Si",
magnetic_flux_density="9.4 T",
rotor_angle="90 deg",
rotor_frequency="14 kHz",
number_of_sidebands=4,
)
Here, lineshape is an instance of the
ShieldingPALineshape class. The required
arguments of this class are the anisotropic_dimension, inverse_dimension, and
channel. We have already defined the first two arguments in the previous
sub-section. The value of the channel argument is the nucleus observed in the MAF
experiment. In this example, this value is ‘29Si’.
The remaining arguments, such as the magnetic_flux_density, rotor_angle,
and rotor_frequency, are set to match the conditions under which the spectrum
was acquired. The value of the number_of_sidebands argument is the number of
sidebands calculated for each line-shape within the kernel.
Once the ShieldingPALineshape instance is created, use the
kernel() method of the
instance to generate the MAF line-shape kernel.
K = lineshape.kernel(supersampling=1)
Data Compression¶
Data compression is optional but recommended. It may reduce the size of the inverse problem and, thus, further computation time.
new_system = TSVDCompression(K, data_object)
compressed_K = new_system.compressed_K
compressed_s = new_system.compressed_s
print(f"truncation_index = {new_system.truncation_index}")
Out:
compression factor = 1.5737704918032787
truncation_index = 61
Solving the inverse problem¶
Smooth-LASSO problem¶
Solve the smooth-lasso problem. You may choose to skip this step and proceed to the statistical learning method. Usually, the statistical learning method is a time-consuming process that solves the smooth-lasso problem over a range of predefined hyperparameters. If you are unsure what range of hyperparameters to use, you can use this step for a quick look into the possible solution by giving a guess value for the \(\alpha\) and \(\lambda\) hyperparameters, and then decide on the hyperparameters range accordingly.
# guess alpha and lambda values.
s_lasso = SmoothLasso(alpha=5e-5, lambda1=5e-6, inverse_dimension=inverse_dimension)
s_lasso.fit(K=compressed_K, s=compressed_s)
f_sol = s_lasso.f
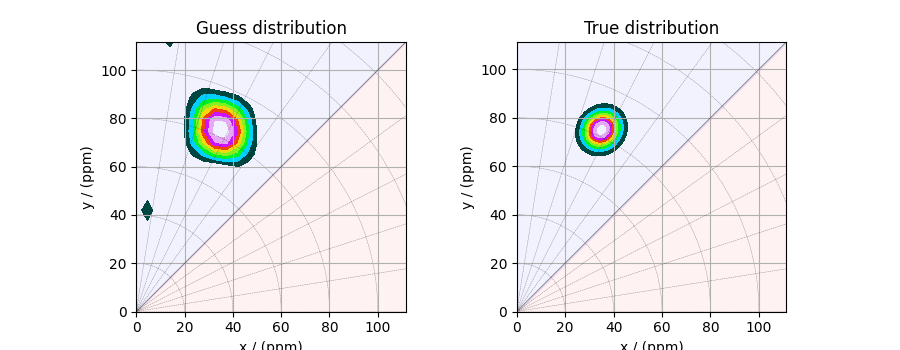
Here, f_sol is the solution corresponding to hyperparameters
\(\alpha=5\times10^{-5}\) and \(\lambda=5\times 10^{-6}\). The plot of this
solution is
_, ax = plt.subplots(1, 2, figsize=(9, 3.5), subplot_kw={"projection": "csdm"})
# the plot of the guess tensor distribution solution.
plot2D(ax[0], f_sol / f_sol.max(), title="Guess distribution")
# the plot of the true tensor distribution.
plot2D(ax[1], true_data_object, title="True distribution")
plt.tight_layout()
plt.show()
Predicted spectrum¶
You may also evaluate the predicted spectrum from the above solution following
residuals = s_lasso.residuals(K, data_object)
predicted_spectrum = data_object - residuals
plt.figure(figsize=(4, 3))
plt.subplot(projection="csdm")
plt.plot(data_object, color="black", label="spectrum") # the original spectrum
plt.plot(predicted_spectrum, color="red", label="prediction") # the predicted spectrum
plt.gca().invert_xaxis()
plt.legend()
plt.tight_layout()
plt.show()
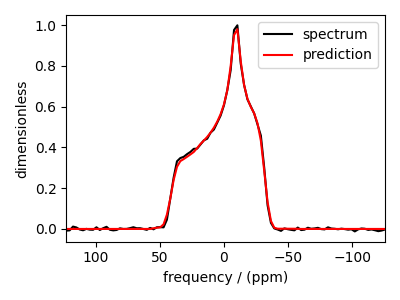
As you can see from the predicted spectrum, our guess isn’t far from the optimum hyperparameters. Let’s create a search grid about the guess hyperparameters and run a cross-validation method for selection.
Statistical learning of the tensors¶
Smooth LASSO cross-validation¶
Create a guess range of values for the \(\alpha\) and \(\lambda\) hyperparameters. The following code generates a range of \(\lambda\) and \(\alpha\) values that are uniformly sampled on the log scale.
lambdas = 10 ** (-5.2 - 1 * (np.arange(6) / 5))
alphas = 10 ** (-4 - 2 * (np.arange(6) / 5))
# set up cross validation smooth lasso method
s_lasso_cv = SmoothLassoCV(
alphas=alphas,
lambdas=lambdas,
inverse_dimension=inverse_dimension,
sigma=0.005,
folds=10,
)
# run the fit using the compressed kernel and compressed signal.
s_lasso_cv.fit(compressed_K, compressed_s)
The optimum hyper-parameters¶
Use the hyperparameters attribute of
the instance for the optimum hyper-parameters, \(\alpha\) and \(\lambda\),
determined from the cross-validation.
print(s_lasso_cv.hyperparameters)
Out:
{'alpha': 6.30957344480193e-06, 'lambda': 1.584893192461114e-06}
The cross-validation surface¶
Optionally, you may want to visualize the cross-validation error curve/surface. Use
the cross_validation_curve attribute
of the instance, as follows. The cross-validation metric is the mean square error
(MSE).
cv_curve = s_lasso_cv.cross_validation_curve
# plot of the cross-validation curve
plt.figure(figsize=(5, 3.5))
ax = plt.subplot(projection="csdm")
ax.contour(np.log10(s_lasso_cv.cross_validation_curve), levels=25)
ax.scatter(
-np.log10(s_lasso_cv.hyperparameters["alpha"]),
-np.log10(s_lasso_cv.hyperparameters["lambda"]),
marker="x",
color="k",
)
plt.tight_layout(pad=0.5)
plt.show()
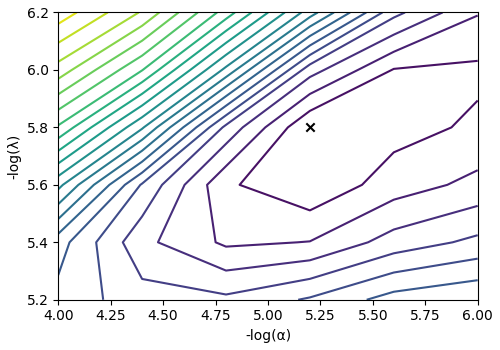
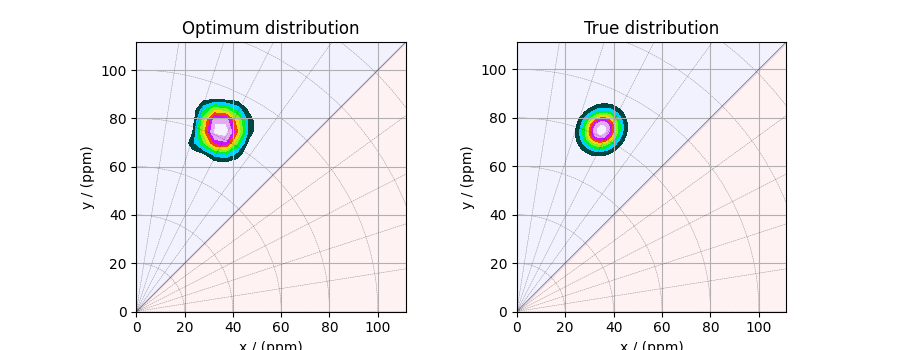
The optimum solution¶
The f attribute of the instance holds
the solution.
The corresponding plot of the solution, along with the true tensor distribution, is shown below.
_, ax = plt.subplots(1, 2, figsize=(9, 3.5), subplot_kw={"projection": "csdm"})
# the plot of the tensor distribution solution.
plot2D(ax[0], f_sol / f_sol.max(), title="Optimum distribution")
# the plot of the true tensor distribution.
plot2D(ax[1], true_data_object, title="True distribution")
plt.tight_layout()
plt.show()
Total running time of the script: ( 0 minutes 19.771 seconds)