from copy import deepcopy
from itertools import combinations
from itertools import product
import csdmpy as cp
import matplotlib.pyplot as plt
import numpy as np
from matplotlib import cm
from mpl_toolkits.mplot3d import Axes3D # lgtm [py/unused-import]
[docs]def to_Haeberlen_grid(csdm_object, zeta, eta, n=5):
"""Convert the three-dimensional p(iso, x, y) to p(iso, zeta, eta) tensor
distribution.
Args
----
csdm_object: CSDM
A CSDM object containing the 3D p(iso, x, y) distribution.
zeta: CSDM.Dimension
A CSDM dimension object describing the zeta dimension.
eta: CSDM.Dimension
A CSDM dimension object describing the eta dimension.
n: int
An integer used in linear interpolation of the data. The default is 5.
"""
[
item.to("ppm", "nmr_frequency_ratio")
for item in csdm_object.x
if item.origin_offset != 0
]
data = csdm_object.y[0].components[0]
extra_dims = 1
if len(csdm_object.x) > 2:
extra_dims = np.sum([item.coordinates.size for item in csdm_object.x[2:]])
data.shape = (extra_dims, data.shape[-2], data.shape[-1])
reg_x, reg_y = (csdm_object.x[i].coordinates.value for i in range(2))
dx = reg_x[1] - reg_x[0]
dy = reg_y[1] - reg_y[0]
sol = np.zeros((extra_dims, zeta.count, eta.count))
bins = [zeta.count, eta.count]
d_zeta = zeta.increment.value / 2
d_eta = eta.increment.value / 2
range_ = [
[zeta.coordinates[0].value - d_zeta, zeta.coordinates[-1].value + d_zeta],
[eta.coordinates[0] - d_eta, eta.coordinates[-1] + d_eta],
]
avg_range_x = (np.arange(n) - (n - 1) / 2) * dx / n
avg_range_y = (np.arange(n) - (n - 1) / 2) * dy / n
for x_item in avg_range_x:
for y_item in avg_range_y:
x__ = np.abs(reg_x + x_item)
y__ = np.abs(reg_y + y_item)
x_, y_ = np.meshgrid(x__, y__)
x_ = x_.ravel()
y_ = y_.ravel()
zeta_grid = np.sqrt(x_**2 + y_**2)
eta_grid = np.ones(zeta_grid.shape)
index = np.where(x_ < y_)
eta_grid[index] = (4 / np.pi) * np.arctan(x_[index] / y_[index])
index = np.where(x_ > y_)
zeta_grid[index] *= -1
eta_grid[index] = (4 / np.pi) * np.arctan(y_[index] / x_[index])
index = np.where(x_ == y_)
np.append(zeta, -zeta_grid[index])
np.append(eta, np.ones(index[0].size))
for i in range(extra_dims):
weight = deepcopy(data[i]).ravel()
weight[index] /= 2
np.append(weight, weight[index])
sol_, _, _ = np.histogram2d(
zeta_grid, eta_grid, weights=weight, bins=bins, range=range_
)
sol[i] += sol_
sol /= n * n
del zeta_grid, eta_grid, index, x_, y_, avg_range_x, avg_range_y
csdm_new = cp.as_csdm(np.squeeze(sol))
csdm_new.x[0] = eta
csdm_new.x[1] = zeta
if len(csdm_object.x) > 2:
csdm_new.x[2] = csdm_object.x[2]
return csdm_new
[docs]def get_polar_grids(ax, ticks=None, offset=0):
"""Generate a piece-wise polar grid of Haeberlen parameters, zeta and eta.
Args:
ax: Matplotlib Axes.
ticks: Tick coordinates where radial grids are drawn. The value can be a list
or a numpy array. The default value is None.
offset: The grid is drawn at an offset away from the origin.
"""
ylim = ax.get_ylim()
xlim = ax.get_xlim()
if ticks is None:
x = np.asarray(ax.get_xticks())
inc = x[1] - x[0]
size = x.size
x = np.arange(size + 5) * inc
else:
x = np.asarray(ticks)
lw = 0.3
t1 = plt.Polygon([[0, 0], [0, x[-1]], [x[-1], x[-1]]], color="b", alpha=0.05)
t2 = plt.Polygon([[0, 0], [x[-1], 0], [x[-1], x[-1]]], color="r", alpha=0.05)
ax.add_artist(t1)
ax.add_artist(t2)
for x_ in x:
if x_ - offset > 0:
ax.add_artist(
plt.Circle(
(0, 0),
x_ - offset,
fill=False,
color="k",
linestyle="--",
linewidth=lw,
alpha=0.5,
)
)
angle1 = np.tan(np.pi * np.asarray([0, 0.2, 0.4, 0.6, 0.8]) / 4.0)
angle2 = np.tan(np.pi * np.asarray([0.8, 0.6, 0.4, 0.2, 0]) / 4.0)
for ang_ in angle1:
ax.plot(x, ((x - offset) * ang_) + offset, "k--", alpha=0.5, linewidth=lw)
for ang_ in angle2:
ax.plot(((x - offset) * ang_) + offset, x, "k--", alpha=0.5, linewidth=lw)
ax.plot(x, x, "k", alpha=0.5, linewidth=2 * lw)
ax.set_xlim(xlim)
ax.set_ylim(ylim)
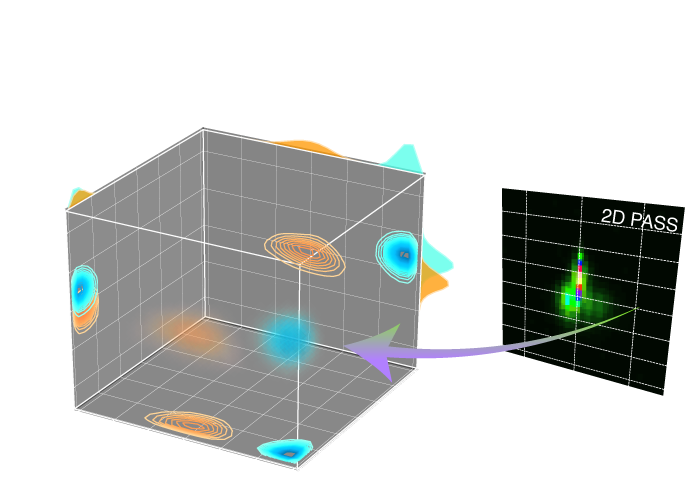
[docs]def plot_3d(
ax,
csdm_objects,
elev=28,
azim=-150,
x_lim=None,
y_lim=None,
z_lim=None,
cmap=cm.PiYG,
box=False,
clip_percent=0.0,
linewidth=1,
alpha=0.15,
**kwargs,
):
r"""Generate a 3D density plot with 2D contour and 1D projections.
Args:
ax: Matplotlib Axes to render the plot.
csdm_objects: A 3D{1} CSDM object or a list of CSDM objects holding the data.
elev: (optional) The 3D view angle, elevation angle in the z plane.
azim: (optional) The 3D view angle, azimuth angle in the x-y plane.
x_lim: (optional) The x limit given as a list, [x_min, x_max].
y_lim: (optional) The y limit given as a list, [y_min, y_max].
z_lim: (optional) The z limit given as a list, [z_min, z_max].
max_2d: (Optional) The normalization factor of the 2D contour projections. The
attribute is meaningful when multiple 3D datasets are viewed on the same
plot. The value is given as a list, [`yz`, `xz`, `xy`], where `ij` is the
maximum of the projection onto the `ij` plane, :math:`i,j \in [x, y, z]`.
max_1d: (Optional) The normalization factor of the 1D projections. The
attribute is meaningful when multiple 3D datasets are viewed on the same
plot. The value is given as a list, [`x`, `y`, `z`], where `i` is the
maximum of the projection onto the `i` axis, :math:`i \in [x, y, z]`.
cmap: (Optional) The colormap or a list of colormaps used in rendering the
volumetric plot. The colormap list is applied to the ordered list of csdm_objects.
The same colormap is used for the 2D contour projections. For 1D plots, the first
color in the colormap scheme is used for the line color.
box: (Optional) If True, draw a box around the 3D data region.
clip_percent: (Optional) The amplitudes of the dataset below the given percent
is made transparent for the volumetric plot.
linewidth: (Optional) The linewidth of the 2D contours, 1D plots and box.
alpha: (Optional) The amount of alpha(transparency) applied in rendering the 3D
volume.
"""
csdm_object_list = csdm_objects
if not isinstance(csdm_objects, list):
csdm_object_list = [csdm_objects]
if not isinstance(cmap, list):
cmap = [cmap]
fraction = np.array([abs(item).sum() for item in csdm_object_list])
fraction /= fraction.max()
for index, item in enumerate(csdm_object_list):
plot_3d_(
ax,
item,
elev=elev,
azim=azim,
x_lim=x_lim,
y_lim=y_lim,
z_lim=z_lim,
fraction=fraction[index],
cmap=cmap[index],
box=box,
clip_percent=clip_percent,
linewidth=linewidth,
alpha=alpha,
**kwargs,
)
def plot_3d_(
ax,
csdm_object,
elev=28,
azim=-150,
x_lim=None,
y_lim=None,
z_lim=None,
fraction=1.0,
cmap=cm.PiYG,
box=False,
clip_percent=0.0,
linewidth=1,
alpha=0.15,
**kwargs,
):
r"""Generate a 3D density plot with 2D contour and 1D projections.
Args:
ax: Matplotlib Axes to render the plot.
csdm_object: A 3D{1} CSDM object holding the data.
elev: (optional) The 3D view angle, elevation angle in the z plane.
azim: (optional) The 3D view angle, azimuth angle in the x-y plane.
x_lim: (optional) The x limit given as a list, [x_min, x_max].
y_lim: (optional) The y limit given as a list, [y_min, y_max].
z_lim: (optional) The z limit given as a list, [z_min, z_max].
max_2d: (Optional) The normalization factor of the 2D contour projections. The
attribute is meaningful when multiple 3D datasets are viewed on the same
plot. The value is given as a list, [`yz`, `xz`, `xy`], where `ij` is the
maximum of the projection onto the `ij` plane, :math:`i,j \in [x, y, z]`.
max_1d: (Optional) The normalization factor of the 1D projections. The
attribute is meaningful when multiple 3D datasets are viewed on the same
plot. The value is given as a list, [`x`, `y`, `z`], where `i` is the
maximum of the projection onto the `i` axis, :math:`i \in [x, y, z]`.
cmap: (Optional) The colormap used in rendering the volumetric plot. The same
colormap is used for the 2D contour projections. For 1D plots, the first
color in the colormap scheme is used for the line color.
box: (Optional) If True, draw a box around the 3D data region.
clip_percent: (Optional) The amplitudes of the dataset below the given percent
is made transparent for the volumetric plot.
linewidth: (Optional) The linewidth of the 2D contours, 1D plots and box.
alpha: (Optional) The amount of alpha(transparency) applied in rendering the 3D
volume.
"""
lw = linewidth
if isinstance(csdm_object, cp.CSDM):
f = csdm_object.y[0].components[0].T
a_, b_, c_ = (item for item in csdm_object.x)
a = a_.coordinates.value
b = b_.coordinates.value
c = c_.coordinates.value
xlabel = f"{a_.axis_label} - 0"
ylabel = f"{b_.axis_label} - 1"
zlabel = f"{c_.axis_label} - 2"
else:
f = csdm_object
a = np.arange(f.shape[0])
b = np.arange(f.shape[1])
c = np.arange(f.shape[2])
xlabel = "x"
ylabel = "y"
zlabel = "z"
delta_a = a[1] - a[0]
delta_b = b[1] - b[0]
delta_c = c[1] - c[0]
clr = cmap
ck = cmap(0)
facecolors = cmap(f)
facecolors[:, :, :, -1] = f * alpha
index = np.where(f < clip_percent / 100)
facecolors[:, :, :, -1][index] = 0
facecolors.shape = (f.size, 4)
if x_lim is None:
x_lim = [a[0], a[-1]]
if y_lim is None:
y_lim = [b[0], b[-1]]
if z_lim is None:
z_lim = [c[0], c[-1]]
offset_scalar = 50
height_scalar = 10
z_offset = (z_lim[1] - z_lim[0]) / offset_scalar
z_scale = np.abs(z_lim[1] - z_lim[0]) / height_scalar
offz_n = z_lim[0] - (delta_c / 2.0)
offz_1d = z_lim[1] + z_offset
y_offset = (y_lim[1] - y_lim[0]) / offset_scalar
y_scale = np.abs(y_lim[1] - y_lim[0]) / height_scalar
offy = y_lim[1] + (delta_b / 2.0)
offy_n_1d = y_lim[0] - y_offset
offx = x_lim[1] + (delta_a / 2.0)
x_scale = np.abs(x_lim[1] - x_lim[0]) / height_scalar
if azim > -90 and azim <= 0:
offx = x_lim[0] - (delta_a / 2.0)
offy_n_1d = y_lim[0] - y_offset
if azim > 0 and azim <= 90:
offy = y_lim[0] - (delta_b / 2.0)
offy_n_1d = y_lim[1] + y_offset
offx = x_lim[0] - (delta_a / 2.0)
if azim > 90:
offx = x_lim[1] + (delta_a / 2.0)
offy_n_1d = y_lim[1] + y_offset
offy = y_lim[0] - (delta_b / 2.0)
ax.set_proj_type("persp")
ax.view_init(elev=elev, azim=azim)
# 2D x-y contour projection ---------------------
levels = (np.arange(20) + 1) / 20
x1, y1 = np.meshgrid(a, b, indexing="ij")
dist_xy = f.mean(axis=2)
dist_xy *= fraction * z_scale / np.abs(dist_xy.max())
ax.contour(
x1,
y1,
dist_xy,
zdir="z",
offset=offz_n,
cmap=clr,
levels=z_scale * levels,
linewidths=lw,
)
# 2D x-z contour projection
x1, y1 = np.meshgrid(a, c, indexing="ij")
dist_xz = f.mean(axis=1)
dist_xz *= fraction * y_scale / np.abs(dist_xz.max())
ax.contour(
x1,
dist_xz,
y1,
zdir="y",
offset=offy,
cmap=clr,
levels=y_scale * levels,
linewidths=lw,
)
# 1D x-axis projection from 2D x-z projection
proj_x = dist_xz.mean(axis=1)
proj_x *= fraction * z_scale / np.abs(proj_x.max())
ax.plot(a, np.sign(z_offset) * proj_x + offz_1d, offy, zdir="y", c=ck, linewidth=lw)
# 1D z-axis projection from 2D x-z projection
proj_z = dist_xz.mean(axis=0)
proj_z *= fraction * y_scale / np.abs(proj_z.max())
ax.plot(
np.sign(azim) * np.sign(y_offset) * proj_z + offy_n_1d,
c,
offx,
zdir="x",
c=ck,
linewidth=lw,
)
ax.set_xlim(z_lim)
# 2D y-z contour projection
x1, y1 = np.meshgrid(b, c, indexing="ij")
dist_yz = f.mean(axis=0)
dist_yz *= fraction * x_scale / np.abs(dist_yz.max())
ax.contour(
dist_yz,
x1,
y1,
zdir="x",
offset=offx,
cmap=clr,
levels=x_scale * levels,
linewidths=lw,
)
# 1D y-axis projection
proj_y = dist_yz.mean(axis=1)
proj_y *= fraction * z_scale / np.abs(proj_y.max())
ax.plot(b, np.sign(z_offset) * proj_y + offz_1d, offx, zdir="x", c=ck, linewidth=lw)
ax.set_xlim(x_lim)
ax.set_ylim(y_lim)
ax.set_zlim(z_lim)
ax.set_xlabel(xlabel)
ax.set_ylabel(ylabel)
ax.set_zlabel(zlabel)
x, y, z = np.meshgrid(a, b, c, indexing="ij")
if "s" not in kwargs:
kwargs["s"] = 300
ax.scatter(x.flat, y.flat, z.flat, marker="X", c=facecolors, **kwargs)
# full box
r1 = [x_lim[0] - delta_a / 2, x_lim[-1] + delta_a / 2]
r2 = [y_lim[0] - delta_b / 2, y_lim[-1] + delta_b / 2]
r3 = [z_lim[-1] - delta_c / 2, z_lim[0] + delta_c / 2]
l_box = lw
for s, e in combinations(np.array(list(product(r1, r2, r3))), 2):
if np.sum(np.abs(s - e)) == r1[1] - r1[0]:
ax.plot3D(*zip(s, e), color="gray", linewidth=l_box)
if np.sum(np.abs(s - e)) == r2[1] - r2[0]:
ax.plot3D(*zip(s, e), color="gray", linewidth=l_box)
if np.sum(np.abs(s - e)) == r3[1] - r3[0]:
ax.plot3D(*zip(s, e), color="gray", linewidth=l_box)
# draw cube
if box:
r1 = [a[0] - delta_a / 2, a[-1] + delta_a / 2]
r2 = [b[0] - delta_b / 2, b[-1] + delta_b / 2]
r3 = [c[0] - delta_c / 2, c[-1] + delta_c / 2]
for s, e in combinations(np.array(list(product(r1, r2, r3))), 2):
if np.sum(np.abs(s - e)) == r1[1] - r1[0]:
ax.plot3D(*zip(s, e), c="blue", linestyle="dashed", linewidth=l_box)
if np.sum(np.abs(s - e)) == r2[1] - r2[0]:
ax.plot3D(*zip(s, e), c="blue", linestyle="dashed", linewidth=l_box)
if np.sum(np.abs(s - e)) == r3[1] - r3[0]:
ax.plot3D(*zip(s, e), c="blue", linestyle="dashed", linewidth=l_box)
ax.xaxis._axinfo["grid"].update({"linewidth": 0.25, "color": "gray"})
ax.yaxis._axinfo["grid"].update({"linewidth": 0.25, "color": "gray"})
ax.zaxis._axinfo["grid"].update({"linewidth": 0.25, "color": "gray"})